-
HTML 5 Document: https://dkfz-odcf.github.io/bioinformatic-reproducibility-course
-
Github Repository: https://github.com/DKFZ-ODCF/bioinformatic-reproducibility-course
Definition "Reproducibility"
Modified after Wikipedia:
Closeness of the agreement between different measurements of the same measurand using the same methodology.
-
Quantified within experiments e.g. as standard deviation
-
Measurements at various scales of mathematical derivation
-
"Raw" data is produced by the sensor hardware
-
-
"… using the same methodology." requires description
-
Interpretability
-
Exact reproducibility from the same raw data
-
|
Note
|
Reproducibility ensures refutability based on methodological reasons! |
Limits of Reproducibility
-
Wet-lab
-
Possible but never exact
-
-
In silico
-
Exact reproducibility seems possible but …
-
… limited by data biases and noise
-
… limited by computer hardware
-
-
|
Note
|
At some point further investment to reproduce results is waste of time! |
Overview
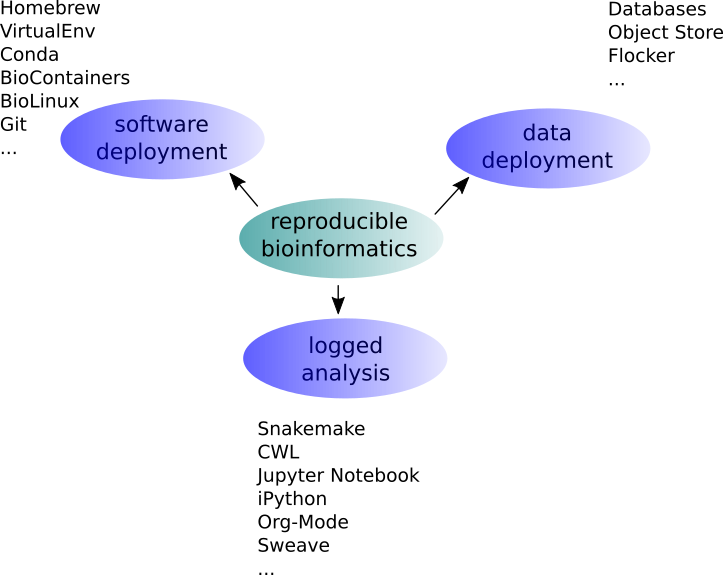
A Bioinformaticians Toolshed
-
Bioinformatic Research Workflow
-
Workflow Management Systems
-
Notes on Programming
-
Version Control
-
Software Deployment, Containers & Virtualiziation
-
Literate Programming
A Bioinformatics Research Workflow
Prototyping
-
Quick & leaky (& correct)
-
Not meant to share
-
Throw away!
-
Aim: Learn about question and implementation
-
Often done in interactive sessions
Exploration
-
Made to last
-
Not meant to share
-
Log analysis, decisions, interpretations
-
Log errors, failure, conclusions & steps towards the correct solution
Reconciliation
-
Based on canonical set of intermediate results
-
Arranged by interpretation & reasoning
-
High certainty and consistency
-
Sufficient to communicate to cooperation partners
Publication
-
Very high certainty
-
Highly tuned towards presentation
-
Selected after reconciliation
-
Publishable document
Workflow Management Systems
-
"workflow" = program
-
Workflow Management Systems
-
Syntax for defining workflows
-
Abstract from execution backends (e.g. different batch processing systems)
-
Manage dependencies between data and processing steps
-
Log execution to ease reproduction
-
Notes on Programming
What is programming?
-
Examples
-
Logging in to a computer checks s.th. on the shell
-
Plotting something in R
-
Composing a workflow in Galaxy
-
Writing a workflow
-
Programming as Communication
-
Computer must understand your code
-
Your future you must understand your code
-
Others must understand your code, because you have to
-
leave the lab
-
explain your approach
-
publish the code
-
Programming as Complexity Management
-
Biological systems are complex
-
Bioinformatic code to analyze biological systems is complex
-
Complexity increases while you add analyses to your project
|
Note
|
Code is living. It changes while you fix bugs and extend it. And it can grow into a monster! |
Programming Languages
-
Every programming language has its strengths and weaknesses
R
| Pro | Con |
|---|---|
|
|
Bash
-
Frequently default shell on Linux environments
| Pro | Con |
|---|---|
|
|
Python, Perl, Ruby or other Scripting Languages
-
Are interpreted and directly executed by an "interpreter"
| Pro | Con |
|---|---|
|
|
Java, C++, C# and other Compiled Languages
-
Translated by a "compiler" into an executible
| Pro | Con |
|---|---|
|
|
|
Note
|
Boundaries are fuzzy: Compilers for scripting languages and scripting features for compiled languages! |
Programming Power Tools
-
Code review
-
Use an integrated development environment [IDE] (PyCharm, IntelliJ IDEA, …)
-
Automated tests
-
Ensure your program remains correct
-
Unit testing frameworks
-
-
Use a version control system
Version Control Systems
-
Manage many versions of your [living] code
-
Code is usually is some form of text and stored in a "repository" (some form of "database")
-
Programming language code (Python, Perl, R, etc.)
-
Workflow descriptions
-
Documentation
-
-
Diverse tools
-
SNV, CVS, Mercurial, Git, …
-
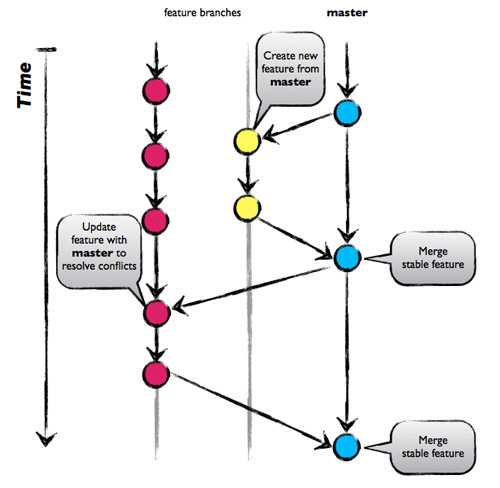
GitHub Flow
-
Commit: registered code version
-
Hash: number associated with commit
-
Branches: parallel development lines
-
Checkout: active registered code base on filesystem (plus uncommitted changes)
-
Master branch: main development line
-
Tag: named commit
How to use?
-
Good and simple guidelines to track development code are:
-
Git Book @ https://git-scm.com/book/en/v2
-
Consider using a Git GUI or an IDE that knows Git
One Step Further
-
Ensure your repo is clean
-
Link data to repository state
-
Put your commit hash into figures and files
-
Git-bindings available for all programming languages
|
Note
|
Tracking code versions is often not enough. Consider using git-lfs. |
R Example with git2r
> library("git2r") # (1)
> repo <- repository("/path/to/your/repo/dir") # (2)
> is_dirty <- function(status) {
length(status$staged) != 0 ||
length(status$unstaged) != 0 ||
length(status$untracked) != 0 # (3)
}
> if (is_dirty(status(repo))) { stop("Not proceeding! Repo is dirty!"); } # (4)
> commitHash <- sha(head(repo)) # (5)-
Load the R library for accessing git repositories
-
Get a handle for the repository
-
Definition of "dirty": there are uncommitted changes or files
-
Check that the repository is clean, i.e. all changes are committed
-
Get the unique identifier of the current repository commit
Python Example with gitpython
---
> from git import * # (1)
> repo = Repo("/path/to/your/repo/dir") # (2)
> if (repo.is_dirty()): raise Exception("Not proceeding! Repo is dirty!") # (3)
> commitHash = repo.head.commit.__str__() # (4)
----
Load the Python library for accessing git repositories
-
Get a handle for the repository
-
Check that the repository is clean, i.e. all changes are committed
-
Get the unique identifier of the current repository commit
Software deployment …
… to publish and share
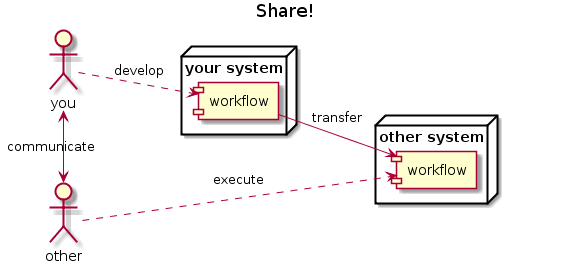
.. to reuse
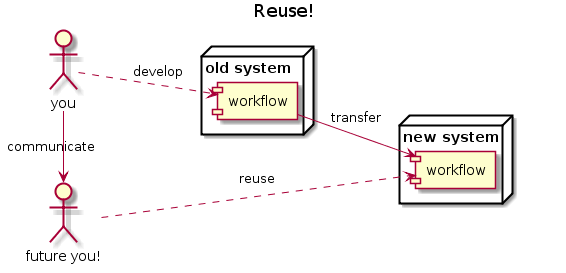
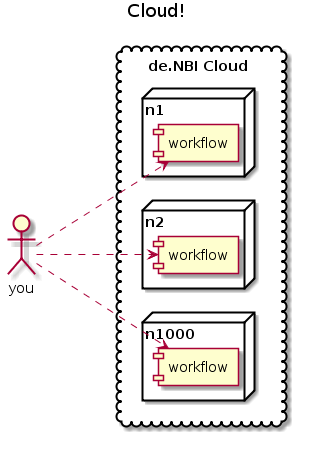
… to scale out
The Challenges
-
Lots of software tools! Lots of versions!
-
Windows, Mac, dozens of Linux distributions, in different versions …
-
Bioinformatic software packages may get lost
-
Do this 1000 times?
-
Boring technical stuff
Packaging System Requirements
-
Quick, easy & correct software deployment
-
Simple user-space installation without administrator rights
-
Manage multiple independent tool sets
-
Lots of packages … maintained by s.b. else ;-D
-
Easy sharing
-
Possible to publish your tools
Enter the realm of Conda
-
Open source software by Anaconda Inc. (Continuum Analytics Inc.)
-
Command-line tool based on Python (2.7, 3.6)
-
Anaconda and Miniconda distributions
-
For Linux > 9000 packages, > 86.000 versions (including those for bioinformatics; June 2018)
-
Linux
-
MacOS
-
Windows
-
… and dive into BioConda
-
Community-driven package repository (channel)
-
> 4.000 bioinformatics related packages, > 18.000 versions
-
BioConda Recipes
-
Most packages available for Linux
-
Final Remarks on Conda
-
Tons of tutorials online
-
Long-term package availability is not 100%
-
Use "bioconda" together with "bioconda-legacy" channel
-
Backup the
pkgs/directory in your Conda installation!
-
Virtualization & Containers
-
Why?
-
You need to scale out to thousands of compute hours
-
Collaboration partners force you to
-
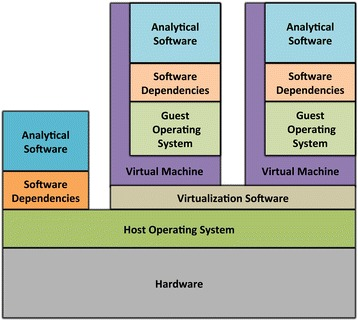
Virtual Machines (VMs)
-
Complete isolation of analysis environment
-
Virtualization software (e.g. also for your desktop)
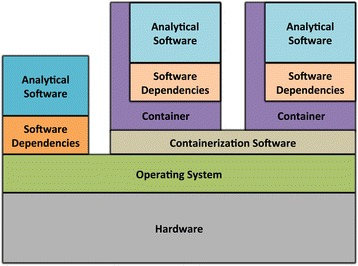
Containers
-
Use host-operating system (kernel)
-
All software and libraries are installed in the container
-
Container technologies
Cloud
-
Usually VMs
-
Simplified handling of multiple VMs
-
start/stop VMs as you need them
-
pay only what you need
-
additional advanced infrastructure at you fingertips
-
Large filesystems on demand
-
Object Store
-
GPUs
-
-
-
Many cloud management systems
-
Commercial
-
Google Cloud, Amazon Web Services, Microsoft Azure, …
-
-
-
Frequently used in science (e.g. de.NBI Cloud)
-
-
-
You need administration knowledge
-
Tools that help you
-
|
Important
|
If you deal with patient related data, public cloud services can be problematic regarding data protection and safety! |
Literate Programming
-
Keep code and documentation together
-
Analysis code
-
Exploratory data analyses
-
Data analysis results and interpretations
-
Decision log
-
Jupyter Notebook
-
Web-server
-
Easy installation via Conda
-
Can run on a large server
-
Can be started with a single command:
jupyter notebook
-
-
Various backends (called "kernels")
-
Bash, Python, R, Spark
-
-
Integrated display code, figures & documentation:
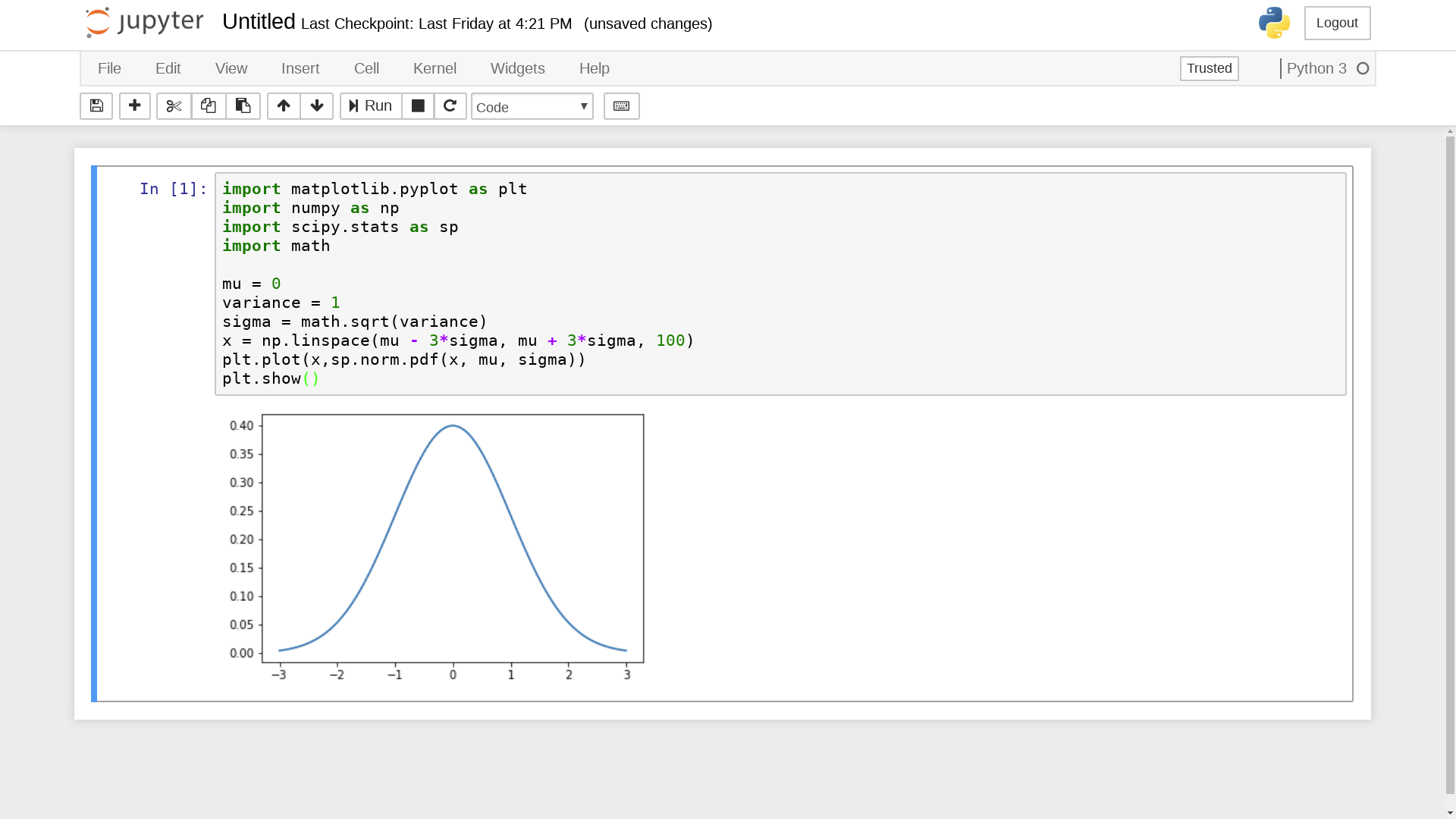
-
Notebooks can be saved and shared
Conclusion
-
Programming is communication & complexity management
-
Bioinformatic reproducibility tools
-
Learnable
-
Doable
-
Worth it!
-
Further Material
-
Courses PM7 and AM4 at the ISMB/ECCB 2019 in Basel @ https://www.iscb.org/ismbeccb2019-program/tutorials
-
Conda User’s Guide @ https://conda.io/docs/user-guide
-
BioConda article @ https://www.nature.com/articles/s41592-018-0046-7
-
NBIS Reproducible Science Course @ https://nbis-reproducible-research.readthedocs.io/en/latest/
-
Source code revisioning with Git
-
Git Book @ https://git-scm.com/book/en/v2
-
Github Flow @ https://guides.github.com/introduction/flow/
-
-
Miniconda @ https://conda.io/miniconda.html
-
BioConda Recipes @ https://github.com/bioconda/bioconda-recipes
-
de.NBI Cloud @ https://www.denbi.de/cloud
References
-
Container & virtualization images by
Piccolo & Framton, Tools and techniques for computational reproducibility, GigaScience, Volume 5, Issue 1, 1 December 2016, s13742-016-0135-4, under Creative Commons Attribution 4.0 International License -
GitFlow image from Nicolas Carlo, 2013
License
Unless otherwise stated, this work and all parts of its are licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.